Discovering critical biomarkers to help match PTSD patients to the most effective treatment
The Biomarker Establishment for Superior Treatment of PTSD Study (BEST)
In partnership with our research collaborators at Stanford University, we studied clinical biomarkers to identify brain activity patterns, or neural signatures, in PTSD within a large Veteran population.
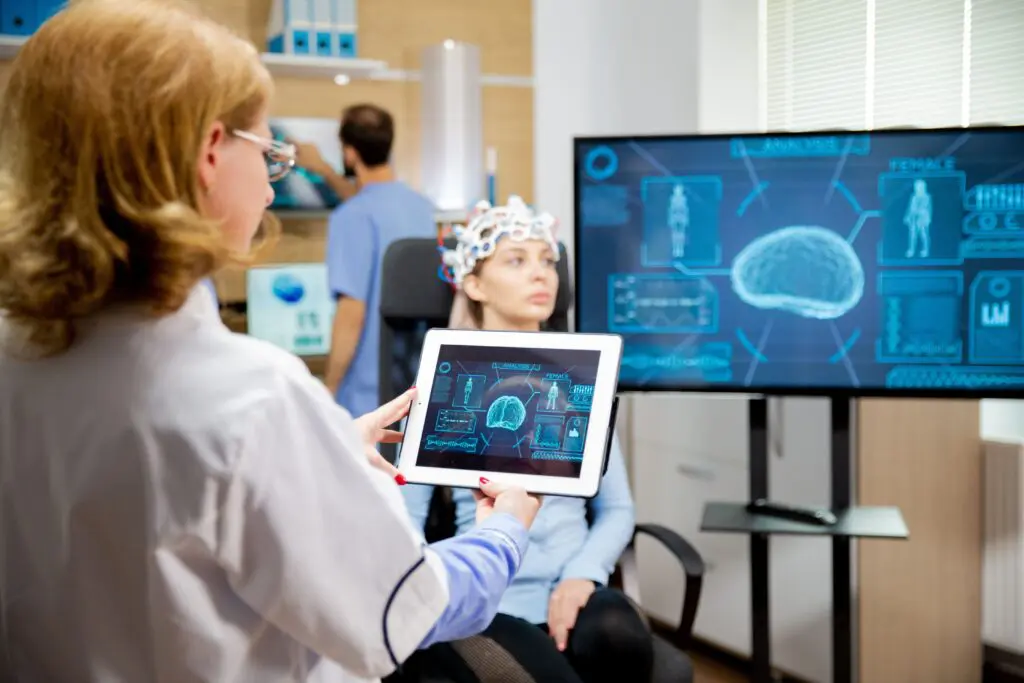
The results showed that PTSD patients could be stratified into groups based on changes in blood flow in a brain circuit, showing a correlation with patients who responded to prolonged exposure psychotherapy, a common PTSD treatment. These findings help move PTSD diagnosis and treatment away from a symptoms-based approach to a biologically-based approach, which can provide the basis for the development of targeted treatments and novel therapeutics.
Our goal is to create a brain signature specific to an individual and then tailor treatment accordingly. Ultimately, we hope to have a clinic-ready tool that will dramatically change how we care for patients with post-traumatic stress disorder.
– Amit Etkin, MD, PhD
Professor of Psychiatry and Behavioral Sciences, Stanford University


Discovering the genetic risk of PTSD
The Global PTSD Genetics Study
Through coordination with more than 200 researchers, across 12 countries, researching genetic data from more than 200,000 people, we demonstrated that the risk of developing PTSD after experiencing trauma is heritable. The study advanced our understanding of PTSD, demonstrating that heritability estimates for PTSD range between 5-20%, which are similar to rates observed for major depression.
We need a drug discovery revolution in PTSD…unbiased genetics approaches will provide the basis for new, rational therapeutics – and may eventually help us better match treatments to patients.
– Karestan C. Koenen PhD
Principal investigator, Stanley Center for Psychiatric Research, Broad Institute and Harvard T.H. Chan School of Public Health

Improving the diagnosis of traumatic brain injury (TBI)
Normative Neuroimaging Library (NNL)
Advanced MRI imaging can now detect microscopic changes in brain structure and/or function caused by trauma or disease. By performing standardized MRI scans on 3,000 adult volunteers who have not been affected by brain trauma or disease, we are forming a library that documents normal population variation. This imaging library would allow physicians to accurately and efficiently diagnose conditions resulting from subtle and early changes in brain structure.

The TBI imaging reference library will provide key information necessary to capitalize on these increasingly available analytical approaches to allow for imaging to help support decisions in the management.
– James R. Stone, MD, PhD
NNL principal investigator, and Vice Chair of Research, UVA Department of Radiology and Medical Imaging


Developing digital health solutions for insomnia
Sleepwell
Sleep disruptions are one of the most debilitating symptoms of PTSD and TBI, and are a risk factor for suicide. We’re developing digital health solutions to validate and analyze data collected from wearables that capture the minute-to-minute experiences of people whose lives are disrupted by PTSD and other brain disorders. Improving methods of measuring sleep will enable the development of precision therapeutics for sleep disorders that are user-friendly, cost-effective, and reliable.
Tracking disease progression in early stage Parkinson’s disease
Parkinson’s Progression Markers Initiative (PPMI) Data Science Modeling Group
The Parkinson’s Progression Markers Initiative (PPMI), sponsored by The Michael J. Fox Foundation (MJFF), is a landmark clinical study designed to identify biomarkers of Parkinson’s disease to predict disease progression and improve patient care. In our aligned missions to advance brain health, MJFF is partnering with CVB’s data science team to quantitatively explore, evaluate, and predict disease trajectory and risk factors using the collected data. The goal of the PPMI is to support the development of new therapies for use in clinical trials.


Technical Performance Verification Study
Sleep disturbance is common in individuals with post-traumatic stress disorder (PTSD). The gold standard assessment for sleep disruptions is the overnight polysomnogram (PSG), which are cumbersome, have a high cost, and not always reflective of how you sleep in your own home. With the growing availability of wearable and home sensor devices that can measure your sleep CVB conducted a study to compare the accuracy of these devices to PSG in people with insomnia and PTSD and (2) understand what each device is measuring and how equivalent their sleep reporting across devices.
Based on rigorous testing in our EarlySignal device lab, we selected the most promising six devices and launched a Technical Performance Verification study with our partners, a leading sleep research team at the University of Pennsylvania. Participants in the study, those with both insomnia and PTSD, spent two nights in the sleep lab wearing the PSG and the six devices in order to help us learn which of these potential sleep devices is the most accurate against the gold standard.
The study completed enrollment in 2022 and results will be updated here and published soon thereafter.
















